| ||||||
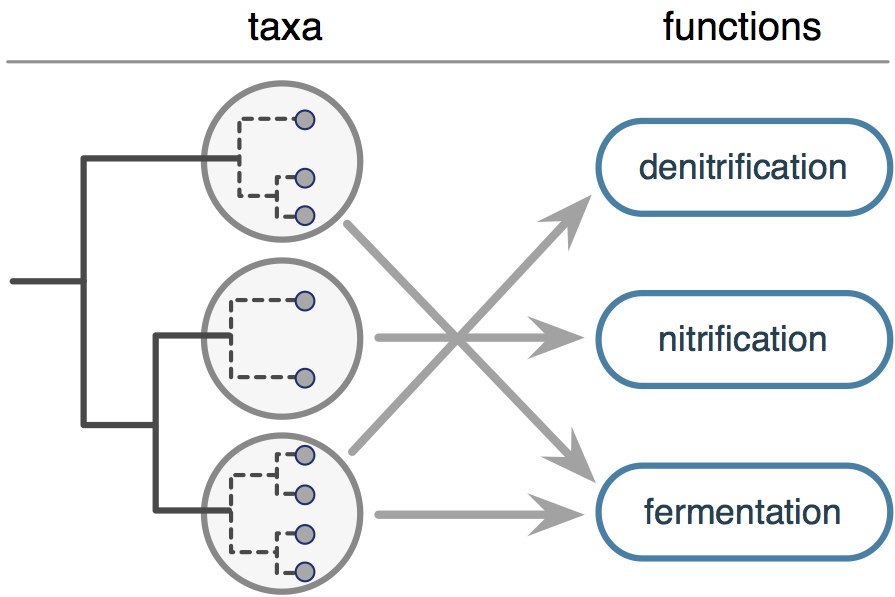
FAPROTAXCode: FAPROTAX01 Predict the likely functional potential of bacterial and archaeal taxa in one or more microbiome samples. The client provides taxonomic identities of queried OTUs and an optional OTU abundance table. High-throughput sequencing of 16S rRNA gene amplicons, also known as 16S metabarcoding, has revealed a vast array of new microbial diversity, with the majority of bacterial and archaeal (aka. prokaryotic) species that are found in natural environments lacking a cultured representative. This means that the metabolic functions and ecological strategies of most detected prokaryotes must be estimated indirectly, for example based on their evolutionary relationships with other better known species. FAPROTAX (Functional Annotation of Prokaryotic Taxa) is a well-established bioinformatics tool for making such inferences, thus facilitating the ecological interpretation of microbiome surveys. FAPROTAX uses a custom database of experimentally confirmed microbial phenotypes, covering over 80 functions of ecological interest, and a custom algorithm for determining the likely functional capabilities of novel prokaryotic taxa. This analysis applies FAPROTAX to a user-provided list of prokaryotic taxa to generate "microbial functional profiles" for your samples.
Caveats and limitations: When using FAPROTAX, keep in mind the following caveats and limitations:
▸ Overview of provided analysis
Our analysis starts with a list of Operational Taxonomic Units (OTUs) with known taxonomic identities, and optionally an OTU abundance table across one or more microbiome samples. Note that your OTUs may represent individual strains, species (most common scenario), genera etc, and FAPROTAX is agnostic with regards to that. We then apply the FAPROTAX annotation pipeline to predict likely functional capabilities for each OTU, and to estimate the overall proportions of various functional groups in your samples. Covered functions mostly cover metabolisms of ecological importance, such as methanogenesis, denitrification, fermentation, photosynthesis etc. Main deliverables:
In addition, we provide a Materials & Methods writeup for use in your publications. ▸ Input requirements
▸ Used 3rd party resources
Main databases and software used in this analysis:
▸ Relevant publications
▸ Price and billing
Price $50 base + $0.01 per OTU. A preview of results is released and payment becomes due upon completion of computations. Payment must be received within 30 days thereafter, and full results become available upon payment.
| ||||||
|
Policies About us Attributions |
Inferentus LLC PO box 554 Pleasant Hill OR, 97455, USA | |||||